refine.bio, Part 2
TL;DR: We need to put together lots of data from disparate sources at low cost via cloud workflows, and this technical post describes the CCDL’s system using specific examples. The refinery runs on AWS and uses Docker and Nomad. Its source code is available on GitHub.
Table of Contents
- Introduction
- The Surveyor System
- The Message System
- The Downloaders System
- The Processors System
- Conclusion
1. Introduction
In refine.bio, Part 1 I explained the need for refine.bio along with many of the specific requirements it would need to meet. In this post I will explain the design I came up to meet those requirements, implementation details about it, and then examples that follow how data is surveyed, downloaded, and then processed.
The design of refine.bio is focused around two abstractions: batches and jobs. A batch is a singular unit of data to be processed. It could be a sample, a genome, or another entity. A job is a unit of work that the system does. There are three kinds of jobs:
- Survey Job: A job that discovers new batches.
- Downloader Job: A job that downloads the data for one or more batches.
- Processor Job: A job that modifies data associated with one or more batches.
The most natural way to discuss the design of refine.bio is in terms of these two abstractions because they represent the core functionality of the system: it refines (via jobs) data (represented by batches). Making these concepts reified entities enables the system to record what it has done and when. The batches table records what data is available, where it can be downloaded from, and any other information the system has about the data. The tables for the various jobs will record when the data was discovered, when it was downloaded, and what processing steps the system performed on it. This will enable a UI to be built exposing this information to users, which will provide them with the context necessary to trust the fidelity of the data.
To help explain how this all comes together I will walk through an example I recently implemented which built Salmon indices for every organism contained in Ensembl Genomes. Salmon is a tool for analyzing RNASeq data which we are using in refine.bio to quantify transcripts. In order to be able to use Salmon, you first need a Salmon index for your transcriptome. Each Salmon index is specific for one organism, so we decided to use refine.bio to preprocess all the indices ahead of time. The basic outline of what we did for each organism is:
- Download the organism’s genome from Ensembl.
- Clean up the genome annotation file.
- Use RSEM to build a custom transcriptome.
- Feed that custom transcriptome into the salmon index command.
We’ll use this diagram as a map to follow our example as the data goes from being stored in Ensembl to being a fully processed Salmon index. I will note here that the code examples used in this post have been stripped down as much as possible to make them easy to read and understand. Therefore almost all of the housekeeping and error handling parts of the code have been omitted along with many comments.
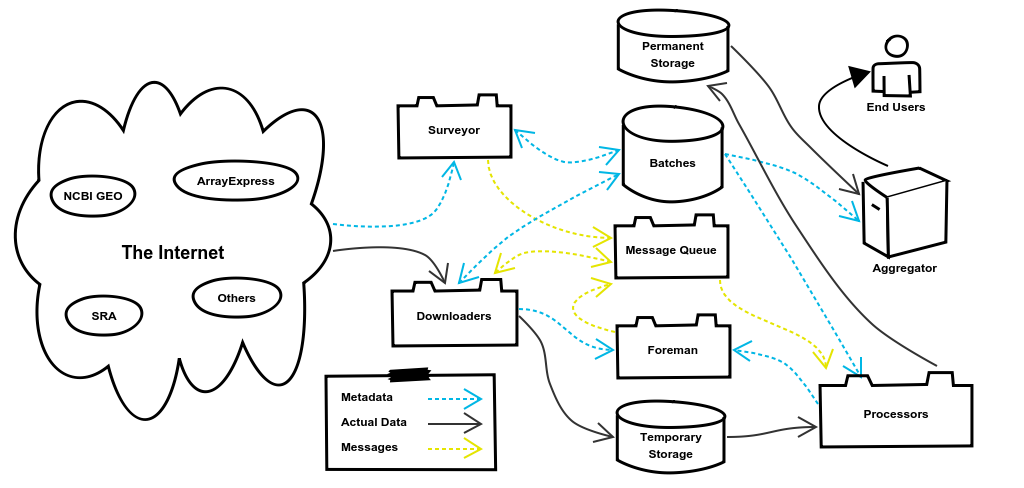
2. The Surveyor System
We’re going to start by looking at how the Surveyor pulls metadata from Ensembl, creates Batches, and then queues DownloaderJobs. The way to create a new Surveyor is to create an implementation of the abstract ExternalSourceSurveyor class. ExternalSourceSurveyorhas three methods which we will need to override:
- discover_batches() - Discovers Batches and add them to the surveyor’s list of Batches by calling theExternalSourceSurveyor.add_batch() method.
- determine_pipeline() - Specifies which Processor Pipeline should run on each Batch. This will be stored as the pipeline_required property on the batches table.
- group_batches() - Groups Batches together that should be downloaded together.
The way these methods will be used together is best demonstrated by looking at the ExternalSourceSurveyor.survey() method:
def survey(self):
self.discover_batches()
for group in self.group_batches():
self.queue_downloader_jobs(group)
So what happens there is that ExternalSourceSurveyor and classes which inherit from it have a property self.batches which is a list of Batches. The discover_batches() method is expected to populate that property via the add_batch() method. For each Batchdiscover_batches() discovers, it calls add_batch(). This function takes required fields for the Batch table, a list of Files associated with the Batch, and a dictionary of key-value pairs which are tacked onto the Batch.
So let’s take a look at what our example’s discover_batches() method looks like:
def discover_batches(self):
ensembl_division = (
SurveyJobKeyValue
.objects
.get(survey_job_id=self.survey_job.id,
key__exact="ensembl_division")
.value
)
r = requests.get(DIVISION_URL_TEMPLATE.format(division=ensembl_division))
# Yes I'm aware that specieses isn't a word. However, I need to
# distinguish between a singular species and multiple species.
specieses = r.json()
for species in specieses:
self._generate_batches(species)
To break down what’s going on: the division of Ensembl which we’re surveying in this particular job is retrieved, a list of species is pulled from Ensembl’s REST API and a Batch is generated for each species. There are quite a few idiosyncrasies when working with the collection of APIs and FTP servers which make up Ensembl Genomes, so I’m not going to delve into everything _generate_batches() does. However, it primarily builds the appropriate URL to download the Batch’s files, creates File objects for both (GTF and FASTA) file types, and cleans up some of the metadata returned by Ensembl’s REST API. However, the most important thing it does is call ExternalSourceSurveyor.add_batch() like so:
self.add_batch(platform_accession_code=platform_accession_code,
experiment_accession_code=url_builder.file_name_species.upper(),
organism_id=url_builder.taxonomy_id,
organism_name=url_builder.scientific_name,
experiment_title="NA",
release_date=current_time,
last_uploaded_date=current_time,
files=[fasta_file, gtf_file],
key_values=species)
add_batch() then takes care of preventing duplicate Batches, creating the Batch objects, setting the correct Processor Pipeline (by calling the user-defined determine_pipeline() method), and saving everything to the database. For this source we only run one Processor Pipeline so we simply return the correct enumerated value. Therefore the implementation of determine_pipeline() is a single line that looks like this:
def determine_pipeline(self, batch: Batch, key_values: Dict = {}) -> ProcessorPipeline:
return ProcessorPipeline.TRANSCRIPTOME_INDEX
At this point we’ve implemented our determine_pipeline() and discover_batches() methods, leaving only group_batches(). Now there’s one detail about the Salmon Indices we’re building that I haven’t yet mentioned: we need two for each organism. We need one that is built to handle samples with a long average transcript read length and one for a short average transcript read length. The way we do this is to have _generate_batches() actually generate two Batches which are identical except that we add a ”kmer_size” key to the key_values dictionary that we pass through to add_batch(). However, both Batches need the same files to run. This setup allows us to only download them once.
We’re able to do this by creating a single DownloaderJob for both Batches. This is exactly what the ExternalSourceSurveyor.group_batches() method is for. We can group the Batches based on the download URL of their Files like so:
def group_batches(self) -> List[List[Batch]]:
"""Groups batches based on the download URL of their first File."""
download_url_mapping = {}
for batch in self.batches:
download_url = batch.files[0].download_url
if download_url in download_url_mapping:
download_url_mapping[download_url].append(batch)
else:
download_url_mapping[download_url] = [batch]
return list(download_url_mapping.values())
If we look back at the ExternalSourceSurveyor.survey() method:
def survey(self):
self.discover_batches()
for group in self.group_batches():
self.queue_downloader_jobs(group)
It will now queue DownloaderJobs for each of the groups returned by the group_batches() method we defined. At this point the Surveyor we’ve implemented will:
- Discover what species are stored in Ensembl.
- Record metadata and create two Batches for each species.
- Queue DownloaderJobs for each group of Batches.
This covers the responsibilities of the Surveyor system. In just a little bit we’ll take a look at what the the Downloaders System does with the jobs we’ve sent it. But first we’ll take a look at how the different systems send messages to each other.
3. The Message System
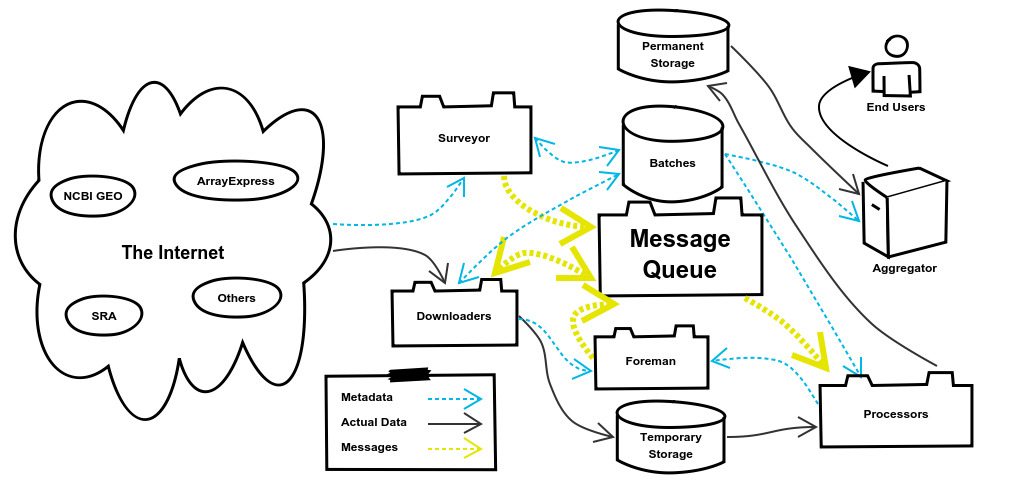
The Message System of refine.bio allows different components to communicate with each other. In the last section I mentioned that the Surveyor System queues DownloaderJobs, but I didn’t define what that meant. Queuing a DownloaderJob has two parts to it. The first creates a database entry representing the DownloaderJob. This entry records information about the job such as when it started, when it ended, and if it was successful. This allows the Foreman to check up on jobs to see if they need to be requeued and is preserved in case it is ever needed to verify the fidelity of a particular Batch of data.
The second part of queuing a DownloaderJob is to send a message to the Downloaders System so it knows that it has work to do. This is how both of these things happen:
@retry(stop_max_attempt_number=3)
def queue_downloader_jobs(self, batches: List[Batch]):
downloader_job = DownloaderJob.create_job_and_relationships(
batches=batches, downloader_task=self.downloader_task())
try:
send_job(downloader_task, downloader_job.id)
except:
# If the task doesn't get sent we don't want the
# downloader_job to be left floating
downloader_job.delete()
raise
The DownloaderJob.create_job_and_relationships() static method creates and saves a DownloaderJob to the database, along with creating links to each of the Batches it should process. Once that is done we send a message to the Downloaders System through the Message System via send_job(). If a failure occurs for any reason, such as a network partition, we delete the DownloaderJob so it’s not left in the database without a message telling the Downloaders System to pick it up. This entire method is wrapped with the decorator @retry(stop_max_attempt_number=3) to attempt to overcome temporary failures.
So, we send a message through the Message System to tell the Downloaders System to pick up the job, but how does that actually work? The Message System is not actually something I’ve built because distributed message queues are a hard problem that a lot of other people have expended a lot of effort to get right. There are a number of tools that could be used for the job such as RabbitMQ, Amazon SQS, or Apache Kafka. We use Hashicorp’s Nomad to manage messages.
Our decision to switch to Nomad was actually unrelated to the Message System. Nomad isn’t actually a message queue but it can be used as one for our purposes. We needed an orchestration tool to manage the cluster we’re going to use for the Downloaders System and the Processors System. Nomad works by running an agent on each node of the cluster. This agent does two primary things: it communicates with the rest of the cluster and it performs fingerprinting on the node. This fingerprinting discovers how many resources the node has available to it such as RAM, disk space, and CPU power. This allows the leader nodes to receive a job message and find a node with the available resources needed to run that job, whether it’s a DownloaderJob or a ProcessorJob. By carefully estimating the amount of resources each job will require, we efficiently use the cluster’s computing power. Nomad’s scheduling system then aims to solve a form of the bin packing problem where jobs are objects with different volumes and the nodes are the bins we want to pack as tightly as possible.
So when I discuss sending a message to the Message System what actually happens is that a Nomad job message is sent to the Nomad leaders, which then find an appropriate node of the cluster to perform the work. However, most contributors to the project won’t have to think too much about what is happening under the hood; they can just send a message to the Message System and trust it to find a node to perform the work associated with that job. So now that we know how the message that our Surveyor sends manages to make it to the Downloaders System we can dive into what that system does.
4. The Downloaders System
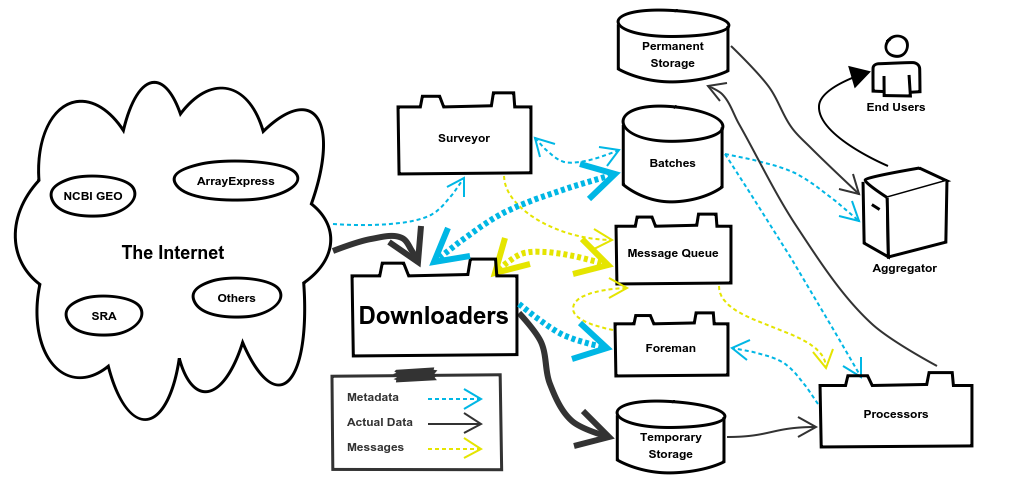
The Downloaders System is responsible for downloading the data for Batches from external sources. Once the data is downloaded it will be stored in the Temporary Storage, which we’re using AWS S3 for. When it comes time to process the data the Processors System, pulls it back out of Temporary Storage. It may seem inefficient to download the data, push it to Temporary Storage, and then pull it back out to process it. Why not just download it and then immediately process it without that other step? There are a couple reasons for this.
If the ProcessorJob fails for any reason we won’t have to re-download the data. Once a DownloaderJob has completed successfully we won’t need to re-download the data from the external source, no matter what issues arise while processing it. Also, this lets us use resources more efficiently. This ties into what I discussed in the last section about how Nomad’s fingerprinting and scheduling capabilities can be used to achieve efficient bin packing. DownloaderJobs have very low resource requirements but can take a while because we can only download data as fast as external servers will provide the data. ProcessorJobs on the other hand tend to have much higher resource requirements because of the processing work we’re performing. If we did not separate these two responsibilities then our MegaJobswould need to be allocated a large number of resources, which they would then not utilize for a long time while downloading the data from external sources. By having ProcessorJobs download the data from within the system, we get the benefit of the good network latency between AWS EC2 and S3.
With your burning questions of “why split those responsibilities?!?!?” answered, we can move on to what a Downloader for our example looks like. I am going to make use of the functions _download_file(download_url: str, file_path: str, job: DownloaderJob) and _upload_files(job_dir: str, files: List[File], job: DownloaderJob) without showing the code for them. This is because they pretty much just do what their names imply; most of their actual implementation involves handling and logging network errors. So here’s the main function for the Downloader:
def download_transcriptome(job_id: int) -> None:
# Block One
job = utils.start_job(job_id)
batches = job.batches.all()
# Prevents different jobs from using the same directory
job_dir = "downloader_job_" + str(job_id)
# Block Two
first_fasta_file = File.objects.get(batch=batches[0], raw_format__exact="fa.gz")
first_gtf_file = File.objects.get(batch=batches[0], raw_format__exact="gtf.gz")
second_fasta_file = File.objects.get(batch=batches[1], raw_format__exact="fa.gz")
second_gtf_file = File.objects.get(batch=batches[1], raw_format__exact="gtf.gz")
os.makedirs(first_fasta_file.get_temp_dir(job_dir), exist_ok=True)
# The two Batches share the same fasta and gtf files, so
# only download each one once
_download_file(first_fasta_file.download_url,
first_fasta_file.get_temp_pre_path(job_dir),
job)
_download_file(first_gtf_file.download_url,
first_gtf_file.get_temp_pre_path(job_dir),
job)
# Block Three
# Then create symlinks so the files for the second Batch
# can be found where they will be expected to.
os.symlink(first_fasta_file.get_temp_pre_path(job_dir),
second_fasta_file.get_temp_pre_path(job_dir))
os.symlink(first_gtf_file.get_temp_pre_path(job_dir),
second_gtf_file.get_temp_pre_path(job_dir))
_upload_files(job_dir,
[first_fasta_file, first_gtf_file, second_fasta_file, second_gtf_file],
job)
utils.end_job(job, batches, success=True)
This function is pretty long and there’re a few different parts so I’ve stuck in a few comments denoting different code blocks. Block One is just getting the job started. It calls utils.start_job() which will mark the job as started and retrieve the DownloaderJob object from the database. We then retrieve the Batches from the database and build our job_dir_prefix which we will put in every path we use during this job so we don’t overlap with any other jobs running concurrently on the same instance. In Block Two we retrieve the File objects out of the database, create a temporary directory to download files into, and then we actually download one set of files.
In Block Three we deal with the fact that we need the same files for two different batches. This goes back to what I discussed in the Surveyor System section about building two Salmon Indices for each organism. Rather than just re-download the exact same files again, we create symlinks to the locations that those files are expected to have been downloaded to. That’s what the File.get_temp_pre_path() method does, it returns where the path where the unprocessed file should be located (pre as in pre-processing). Now when we call the _upload_files() function it will have each of the Files we passed into it upload themselves. That’s right, each File object is capable of uploading itself to Temporary Storage via its upload_raw_file() method. Finally we just call utils.end_job() to mark the job as completed successfully. (In the actual function we catch errors and call utils.end_job() with success=False when appropriate.)
The one other thing that utils.end_job() will take care of for us is queuing a ProcessorJob. This is very similar to the way a DownloaderJob was queued earlier, so I’ll just move along to…
5. The Processors System
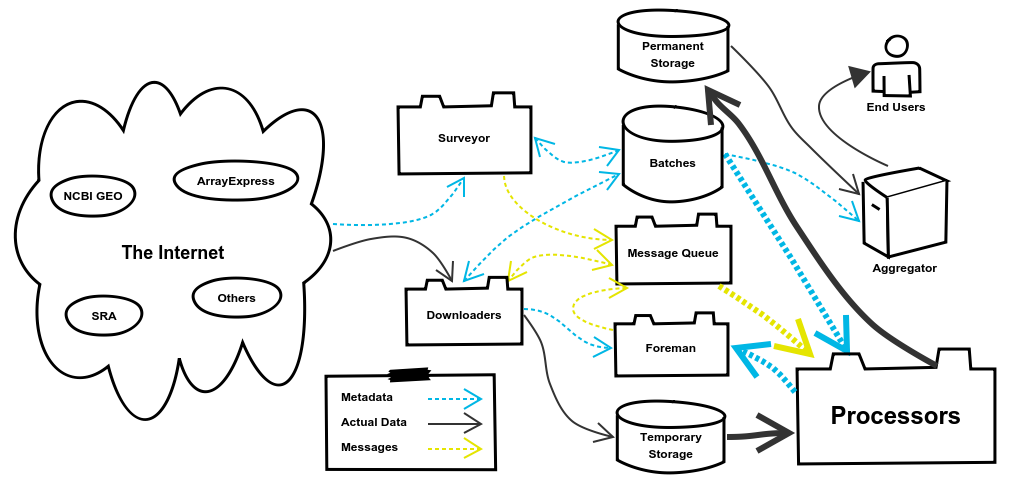
You’ve made it to the good stuff! The Processors System puts the refine in refine.bio. The full range of types of processing that we actually perform is outside the scope of this blog post, but they are carefully considered by the project’s Scientific Lead, Dr. Jaclyn Taroni. Instead we’ll focus on just the example we’ve been following throughout this post. The basic flow for all Processors is to retrieve the Batch’s data from Temporary Storage, perform some kind of processing, and then push the processed data into Permanent Storage. The processing we’ll need to perform for our example are the steps I outlined in the beginning of the post:
- Clean the genome annotation file.
- Use RSEM to build an custom transcriptome.
- Feed that custom transcriptome into the salmon index command.
However, before we can dive into actually doing all of that we need to discuss what Processors are. A Processor is actually just a function that accepts a single argument: the ID of a ProcessorJob. When the Processor System receives a job message from the Message System it will extract the ProcessorJob ID from the message along with the name of the Processor to execute. It then runs the function and waits for it to complete so it can report that the job’s computational resources can be used by other jobs.
Despite the minimal specification of Processors, there are certain responsibilities they are expected to complete. Each Processor must mark when it starts the job, when it ends the job, and whether or not the job was successful. They should also free up any disk space they utilize and if they’re successful they should remove the raw data from Temporary Storage. Since every Processor needs to do this stuff a framework is provided by the system to make it easy and to keep things DRY. The framework was designed with the goal of allowing reusable and composable functions to be created which can then be joined together into a single pipeline. Each function in the pipeline takes a job_context dictionary as an input and must output another (or the same) dictionary which can be passed into the next function of the pipeline. This behavior can be utilized by calling processors.utils.run_pipeline(), which takes two arguments: an initial job_context dictionary and a list of functions.
To illustrate this let’s take a look at the Processor function for our example:
def build_index(job_id: int) -> None:
utils.run_pipeline({"job_id": job_id},
[utils.start_job,
_set_job_prefix,
_prepare_files,
_process_gtf,
_create_index,
_zip_index,
utils.upload_processed_files,
utils.cleanup_raw_files,
utils.end_job])
Any function which returns a job_context containing ”success”: False will cause the pipeline to terminate and the job to be marked as a failure. Notice that the list of functions is composed of calls to functions defined in the utils namespace and private functions from the current namespace. The functions defined in the utils namespace take care of the common functionality I discussed in the previous paragraph. They pretty much all do what their names imply, but it will be helpful to note that in addition to marking the ProcessorJob as started, utils.start_job() also retrieves the ProcessorJob and Batch objects from the database and adds them to the job_context. So now let’s take a look at our pipeline step by step starting with:
def _set_job_prefix(job_context: Dict) -> Dict:
job_context["job_dir_prefix"] = "processor_job_" + str(job_context["job_id"])
return job_context
This adds the key “job_dir_prefix” to the job_context, which serves a similar purpose to the variable named similarly from the Downloaders System section: it makes all paths related to this job unique on the local disk. Next we run _prepare_files() which has the purpose of pulling the files from Temporary Storage and unzipping them so they’re ready to be processed.
def _prepare_files(job_context: Dict) -> Dict:
# Transcriptome index processor jobs have only one batch. Each
# batch has a fasta file and a gtf file
batch = job_context["batches"][0]
fasta_file = File.objects.get(batch=batch, raw_format__exact="fa.gz")
gtf_file = File.objects.get(batch=batch, raw_format__exact="gtf.gz")
fasta_file.download_raw_file(job_context["job_dir_prefix"])
gtf_file.download_raw_file(job_context["job_dir_prefix"])
gzipped_fasta_file_path = fasta_file.get_temp_pre_path(job_context["job_dir_prefix"])
gzipped_gtf_file_path = gtf_file.get_temp_pre_path(job_context["job_dir_prefix"])
job_context["fasta_file_path"] = gzipped_fasta_file_path.replace(".gz", "")
job_context["gtf_file_path"] = gzipped_gtf_file_path.replace(".gz", "")
with gzip.open(gzipped_fasta_file_path, "rb") as gzipped_file, \
open(job_context["fasta_file_path"], "wb") as gunzipped_file:
shutil.copyfileobj(gzipped_file, gunzipped_file)
with gzip.open(gzipped_gtf_file_path, "rb") as gzipped_file, \
open(job_context["gtf_file_path"], "wb") as gunzipped_file:
shutil.copyfileobj(gzipped_file, gunzipped_file)
job_context["fasta_file"] = fasta_file
job_context["gtf_file"] = gtf_file
return job_context
With our files unzipped locally, we’re ready to start processing by calling process_gtf(). This function cleans the genome annotation file (the gtf_file in this case) and creates a mapping from genes to transcripts for the given genome. If you’re not sure what that means, don’t worry about it; the upshot is that job_context["gtf_file_path"] is updated to point to the cleaned GTF file and the ”genes_to_transcripts_path” key is added to the job_context. Now we’re ready to create a Salmon Index using RSEM and Salmon:
def _create_index(job_context: Dict) -> Dict:
# Prepare the output directories.
work_dir = job_context["gtf_file"].get_temp_dir(job_context["job_dir_prefix"])
job_context["output_dir"] = os.path.join(work_dir, "index")
rsem_index_dir = os.path.join(work_dir, "rsem_index")
os.makedirs(rsem_index_dir, exist_ok=True)
# RSEM takes a prefix path and then all files generated by it will
# start with that
rsem_prefix = os.path.join(rsem_index_dir,
job_context["fasta_file"].get_base_name())
rsem_command_string = (
"rsem-prepare-reference --gtf {gtf_file}"
" --transcript-to-gene-map {genes_to_transcripts} {fasta_file} {rsem_prefix}"
)
rsem_formatted_command = rsem_command_string.format(
gtf_file=job_context["gtf_file_path"],
genes_to_transcripts=job_context["genes_to_transcripts_path"],
fasta_file=job_context["fasta_file_path"],
rsem_prefix=rsem_prefix
)
rsem_completed_command = subprocess.run(rsem_formatted_command.split())
salmon_command_string = ("salmon --no-version-check index -t {rsem_transcripts}"
" -i {index_dir} --type quasi -k {kmer_size}")
# This is the property we added in the Surveyor System section
# to denote whether this index is for long or short average read lengths.
kmer_size_property = BatchKeyValue.objects.get(batch=job_context["batches"][0],
key="kmer_size")
# rsem-prepare-reference outputs a transcripts.fa file which needs
# to be passed into salmon.
rsem_transcripts = rsem_prefix + ".transcripts.fa"
salmon_formatted_command = salmon_command_string.format(
rsem_transcripts=rsem_transcripts,
index_dir=job_context["output_dir"],
kmer_size=kmer_size_property.value)
salmon_completed_command = subprocess.run(salmon_formatted_command.split())
return job_context
It’s a big function, perhaps even a bit too big, but it really boils down to two shell calls. The first shell call to rsem-prepare-referenceoutputs the rsem_transcripts file which is then passed into salmon index. salmon-index outputs an entire directory which we want all of.
The next call in the pipeline, _zip_index(), takes care of zipping the entire directory into a single tarball along with setting job_context["files_to_upload"] = [job_context["gtf_file"]]. The reason for setting that property is that utils.upload_processed_files will look for it and, if it’s present in job_context, will only upload the files in that list to Permanent Storage. The File.upload_processed_file() method will use the File’s processed_format property to determine which file should be uploaded. Therefore even though we don’t want to upload the gtf_file, that File object knows how to upload its processed file, which in this case is the tarball we create in _zip_index(). All that’s left in our pipeline is utils.cleanup_raw_files() which will clear the raw data out of Temporary Storage and utils.end_job() which will free up the local disk space for the next job and mark the job as a success.
6. Conclusion
Once the ProcessorJob has been marked as a success in the database, we’re done with our example! We’ve certainly skipped over some sticky parts such as dealing with the idiosyncrasies of Ensembl’s REST API or less interesting code such as failure logging and handling. However, the goal of this blog post was to illustrate how the different components of the system work together to actually get some data from The Internet™ and refine it. I also only barely mentioned the Foreman, which makes sure that when failure inevitably happens, jobs will be retried a couple times before an administrator of the system is notified.
I’ve also omitted a discussion of the Aggregator, but that’s mostly because it’s still being fully specced out. What is defined about it is that it will have a REST API which our various interfaces will use to search, aggregate, and download data stored in refine.bio’s compendium. We’re currently gathering user feedback to better define its functionality. We’re also currently working on finding additional team members to help us implement it.
TL;DR: We need to put together lots of data from disparate sources at low cost via cloud workflows, and this technical post describes the CCDL’s system using specific examples. The refinery runs on AWS and uses Docker and Nomad. Its source code is available on GitHub.
Table of Contents
- Introduction
- The Surveyor System
- The Message System
- The Downloaders System
- The Processors System
- Conclusion
1. Introduction
In refine.bio, Part 1 I explained the need for refine.bio along with many of the specific requirements it would need to meet. In this post I will explain the design I came up to meet those requirements, implementation details about it, and then examples that follow how data is surveyed, downloaded, and then processed.
The design of refine.bio is focused around two abstractions: batches and jobs. A batch is a singular unit of data to be processed. It could be a sample, a genome, or another entity. A job is a unit of work that the system does. There are three kinds of jobs:
- Survey Job: A job that discovers new batches.
- Downloader Job: A job that downloads the data for one or more batches.
- Processor Job: A job that modifies data associated with one or more batches.
The most natural way to discuss the design of refine.bio is in terms of these two abstractions because they represent the core functionality of the system: it refines (via jobs) data (represented by batches). Making these concepts reified entities enables the system to record what it has done and when. The batches table records what data is available, where it can be downloaded from, and any other information the system has about the data. The tables for the various jobs will record when the data was discovered, when it was downloaded, and what processing steps the system performed on it. This will enable a UI to be built exposing this information to users, which will provide them with the context necessary to trust the fidelity of the data.
To help explain how this all comes together I will walk through an example I recently implemented which built Salmon indices for every organism contained in Ensembl Genomes. Salmon is a tool for analyzing RNASeq data which we are using in refine.bio to quantify transcripts. In order to be able to use Salmon, you first need a Salmon index for your transcriptome. Each Salmon index is specific for one organism, so we decided to use refine.bio to preprocess all the indices ahead of time. The basic outline of what we did for each organism is:
- Download the organism’s genome from Ensembl.
- Clean up the genome annotation file.
- Use RSEM to build a custom transcriptome.
- Feed that custom transcriptome into the salmon index command.
We’ll use this diagram as a map to follow our example as the data goes from being stored in Ensembl to being a fully processed Salmon index. I will note here that the code examples used in this post have been stripped down as much as possible to make them easy to read and understand. Therefore almost all of the housekeeping and error handling parts of the code have been omitted along with many comments.

2. The Surveyor System
We’re going to start by looking at how the Surveyor pulls metadata from Ensembl, creates Batches, and then queues DownloaderJobs. The way to create a new Surveyor is to create an implementation of the abstract ExternalSourceSurveyor class. ExternalSourceSurveyorhas three methods which we will need to override:
- discover_batches() - Discovers Batches and add them to the surveyor’s list of Batches by calling theExternalSourceSurveyor.add_batch() method.
- determine_pipeline() - Specifies which Processor Pipeline should run on each Batch. This will be stored as the pipeline_required property on the batches table.
- group_batches() - Groups Batches together that should be downloaded together.
The way these methods will be used together is best demonstrated by looking at the ExternalSourceSurveyor.survey() method:
def survey(self):
self.discover_batches()
for group in self.group_batches():
self.queue_downloader_jobs(group)
So what happens there is that ExternalSourceSurveyor and classes which inherit from it have a property self.batches which is a list of Batches. The discover_batches() method is expected to populate that property via the add_batch() method. For each Batchdiscover_batches() discovers, it calls add_batch(). This function takes required fields for the Batch table, a list of Files associated with the Batch, and a dictionary of key-value pairs which are tacked onto the Batch.
So let’s take a look at what our example’s discover_batches() method looks like:
def discover_batches(self):
ensembl_division = (
SurveyJobKeyValue
.objects
.get(survey_job_id=self.survey_job.id,
key__exact="ensembl_division")
.value
)
r = requests.get(DIVISION_URL_TEMPLATE.format(division=ensembl_division))
# Yes I'm aware that specieses isn't a word. However, I need to
# distinguish between a singular species and multiple species.
specieses = r.json()
for species in specieses:
self._generate_batches(species)
To break down what’s going on: the division of Ensembl which we’re surveying in this particular job is retrieved, a list of species is pulled from Ensembl’s REST API and a Batch is generated for each species. There are quite a few idiosyncrasies when working with the collection of APIs and FTP servers which make up Ensembl Genomes, so I’m not going to delve into everything _generate_batches() does. However, it primarily builds the appropriate URL to download the Batch’s files, creates File objects for both (GTF and FASTA) file types, and cleans up some of the metadata returned by Ensembl’s REST API. However, the most important thing it does is call ExternalSourceSurveyor.add_batch() like so:
self.add_batch(platform_accession_code=platform_accession_code,
experiment_accession_code=url_builder.file_name_species.upper(),
organism_id=url_builder.taxonomy_id,
organism_name=url_builder.scientific_name,
experiment_title="NA",
release_date=current_time,
last_uploaded_date=current_time,
files=[fasta_file, gtf_file],
key_values=species)
add_batch() then takes care of preventing duplicate Batches, creating the Batch objects, setting the correct Processor Pipeline (by calling the user-defined determine_pipeline() method), and saving everything to the database. For this source we only run one Processor Pipeline so we simply return the correct enumerated value. Therefore the implementation of determine_pipeline() is a single line that looks like this:
def determine_pipeline(self, batch: Batch, key_values: Dict = {}) -> ProcessorPipeline:
return ProcessorPipeline.TRANSCRIPTOME_INDEX
At this point we’ve implemented our determine_pipeline() and discover_batches() methods, leaving only group_batches(). Now there’s one detail about the Salmon Indices we’re building that I haven’t yet mentioned: we need two for each organism. We need one that is built to handle samples with a long average transcript read length and one for a short average transcript read length. The way we do this is to have _generate_batches() actually generate two Batches which are identical except that we add a ”kmer_size” key to the key_values dictionary that we pass through to add_batch(). However, both Batches need the same files to run. This setup allows us to only download them once.
We’re able to do this by creating a single DownloaderJob for both Batches. This is exactly what the ExternalSourceSurveyor.group_batches() method is for. We can group the Batches based on the download URL of their Files like so:
def group_batches(self) -> List[List[Batch]]:
"""Groups batches based on the download URL of their first File."""
download_url_mapping = {}
for batch in self.batches:
download_url = batch.files[0].download_url
if download_url in download_url_mapping:
download_url_mapping[download_url].append(batch)
else:
download_url_mapping[download_url] = [batch]
return list(download_url_mapping.values())
If we look back at the ExternalSourceSurveyor.survey() method:
def survey(self):
self.discover_batches()
for group in self.group_batches():
self.queue_downloader_jobs(group)
It will now queue DownloaderJobs for each of the groups returned by the group_batches() method we defined. At this point the Surveyor we’ve implemented will:
- Discover what species are stored in Ensembl.
- Record metadata and create two Batches for each species.
- Queue DownloaderJobs for each group of Batches.
This covers the responsibilities of the Surveyor system. In just a little bit we’ll take a look at what the the Downloaders System does with the jobs we’ve sent it. But first we’ll take a look at how the different systems send messages to each other.
3. The Message System

The Message System of refine.bio allows different components to communicate with each other. In the last section I mentioned that the Surveyor System queues DownloaderJobs, but I didn’t define what that meant. Queuing a DownloaderJob has two parts to it. The first creates a database entry representing the DownloaderJob. This entry records information about the job such as when it started, when it ended, and if it was successful. This allows the Foreman to check up on jobs to see if they need to be requeued and is preserved in case it is ever needed to verify the fidelity of a particular Batch of data.
The second part of queuing a DownloaderJob is to send a message to the Downloaders System so it knows that it has work to do. This is how both of these things happen:
@retry(stop_max_attempt_number=3)
def queue_downloader_jobs(self, batches: List[Batch]):
downloader_job = DownloaderJob.create_job_and_relationships(
batches=batches, downloader_task=self.downloader_task())
try:
send_job(downloader_task, downloader_job.id)
except:
# If the task doesn't get sent we don't want the
# downloader_job to be left floating
downloader_job.delete()
raise
The DownloaderJob.create_job_and_relationships() static method creates and saves a DownloaderJob to the database, along with creating links to each of the Batches it should process. Once that is done we send a message to the Downloaders System through the Message System via send_job(). If a failure occurs for any reason, such as a network partition, we delete the DownloaderJob so it’s not left in the database without a message telling the Downloaders System to pick it up. This entire method is wrapped with the decorator @retry(stop_max_attempt_number=3) to attempt to overcome temporary failures.
So, we send a message through the Message System to tell the Downloaders System to pick up the job, but how does that actually work? The Message System is not actually something I’ve built because distributed message queues are a hard problem that a lot of other people have expended a lot of effort to get right. There are a number of tools that could be used for the job such as RabbitMQ, Amazon SQS, or Apache Kafka. We use Hashicorp’s Nomad to manage messages.
Our decision to switch to Nomad was actually unrelated to the Message System. Nomad isn’t actually a message queue but it can be used as one for our purposes. We needed an orchestration tool to manage the cluster we’re going to use for the Downloaders System and the Processors System. Nomad works by running an agent on each node of the cluster. This agent does two primary things: it communicates with the rest of the cluster and it performs fingerprinting on the node. This fingerprinting discovers how many resources the node has available to it such as RAM, disk space, and CPU power. This allows the leader nodes to receive a job message and find a node with the available resources needed to run that job, whether it’s a DownloaderJob or a ProcessorJob. By carefully estimating the amount of resources each job will require, we efficiently use the cluster’s computing power. Nomad’s scheduling system then aims to solve a form of the bin packing problem where jobs are objects with different volumes and the nodes are the bins we want to pack as tightly as possible.
So when I discuss sending a message to the Message System what actually happens is that a Nomad job message is sent to the Nomad leaders, which then find an appropriate node of the cluster to perform the work. However, most contributors to the project won’t have to think too much about what is happening under the hood; they can just send a message to the Message System and trust it to find a node to perform the work associated with that job. So now that we know how the message that our Surveyor sends manages to make it to the Downloaders System we can dive into what that system does.
4. The Downloaders System

The Downloaders System is responsible for downloading the data for Batches from external sources. Once the data is downloaded it will be stored in the Temporary Storage, which we’re using AWS S3 for. When it comes time to process the data the Processors System, pulls it back out of Temporary Storage. It may seem inefficient to download the data, push it to Temporary Storage, and then pull it back out to process it. Why not just download it and then immediately process it without that other step? There are a couple reasons for this.
If the ProcessorJob fails for any reason we won’t have to re-download the data. Once a DownloaderJob has completed successfully we won’t need to re-download the data from the external source, no matter what issues arise while processing it. Also, this lets us use resources more efficiently. This ties into what I discussed in the last section about how Nomad’s fingerprinting and scheduling capabilities can be used to achieve efficient bin packing. DownloaderJobs have very low resource requirements but can take a while because we can only download data as fast as external servers will provide the data. ProcessorJobs on the other hand tend to have much higher resource requirements because of the processing work we’re performing. If we did not separate these two responsibilities then our MegaJobswould need to be allocated a large number of resources, which they would then not utilize for a long time while downloading the data from external sources. By having ProcessorJobs download the data from within the system, we get the benefit of the good network latency between AWS EC2 and S3.
With your burning questions of “why split those responsibilities?!?!?” answered, we can move on to what a Downloader for our example looks like. I am going to make use of the functions _download_file(download_url: str, file_path: str, job: DownloaderJob) and _upload_files(job_dir: str, files: List[File], job: DownloaderJob) without showing the code for them. This is because they pretty much just do what their names imply; most of their actual implementation involves handling and logging network errors. So here’s the main function for the Downloader:
def download_transcriptome(job_id: int) -> None:
# Block One
job = utils.start_job(job_id)
batches = job.batches.all()
# Prevents different jobs from using the same directory
job_dir = "downloader_job_" + str(job_id)
# Block Two
first_fasta_file = File.objects.get(batch=batches[0], raw_format__exact="fa.gz")
first_gtf_file = File.objects.get(batch=batches[0], raw_format__exact="gtf.gz")
second_fasta_file = File.objects.get(batch=batches[1], raw_format__exact="fa.gz")
second_gtf_file = File.objects.get(batch=batches[1], raw_format__exact="gtf.gz")
os.makedirs(first_fasta_file.get_temp_dir(job_dir), exist_ok=True)
# The two Batches share the same fasta and gtf files, so
# only download each one once
_download_file(first_fasta_file.download_url,
first_fasta_file.get_temp_pre_path(job_dir),
job)
_download_file(first_gtf_file.download_url,
first_gtf_file.get_temp_pre_path(job_dir),
job)
# Block Three
# Then create symlinks so the files for the second Batch
# can be found where they will be expected to.
os.symlink(first_fasta_file.get_temp_pre_path(job_dir),
second_fasta_file.get_temp_pre_path(job_dir))
os.symlink(first_gtf_file.get_temp_pre_path(job_dir),
second_gtf_file.get_temp_pre_path(job_dir))
_upload_files(job_dir,
[first_fasta_file, first_gtf_file, second_fasta_file, second_gtf_file],
job)
utils.end_job(job, batches, success=True)
This function is pretty long and there’re a few different parts so I’ve stuck in a few comments denoting different code blocks. Block One is just getting the job started. It calls utils.start_job() which will mark the job as started and retrieve the DownloaderJob object from the database. We then retrieve the Batches from the database and build our job_dir_prefix which we will put in every path we use during this job so we don’t overlap with any other jobs running concurrently on the same instance. In Block Two we retrieve the File objects out of the database, create a temporary directory to download files into, and then we actually download one set of files.
In Block Three we deal with the fact that we need the same files for two different batches. This goes back to what I discussed in the Surveyor System section about building two Salmon Indices for each organism. Rather than just re-download the exact same files again, we create symlinks to the locations that those files are expected to have been downloaded to. That’s what the File.get_temp_pre_path() method does, it returns where the path where the unprocessed file should be located (pre as in pre-processing). Now when we call the _upload_files() function it will have each of the Files we passed into it upload themselves. That’s right, each File object is capable of uploading itself to Temporary Storage via its upload_raw_file() method. Finally we just call utils.end_job() to mark the job as completed successfully. (In the actual function we catch errors and call utils.end_job() with success=False when appropriate.)
The one other thing that utils.end_job() will take care of for us is queuing a ProcessorJob. This is very similar to the way a DownloaderJob was queued earlier, so I’ll just move along to…
5. The Processors System

You’ve made it to the good stuff! The Processors System puts the refine in refine.bio. The full range of types of processing that we actually perform is outside the scope of this blog post, but they are carefully considered by the project’s Scientific Lead, Dr. Jaclyn Taroni. Instead we’ll focus on just the example we’ve been following throughout this post. The basic flow for all Processors is to retrieve the Batch’s data from Temporary Storage, perform some kind of processing, and then push the processed data into Permanent Storage. The processing we’ll need to perform for our example are the steps I outlined in the beginning of the post:
- Clean the genome annotation file.
- Use RSEM to build an custom transcriptome.
- Feed that custom transcriptome into the salmon index command.
However, before we can dive into actually doing all of that we need to discuss what Processors are. A Processor is actually just a function that accepts a single argument: the ID of a ProcessorJob. When the Processor System receives a job message from the Message System it will extract the ProcessorJob ID from the message along with the name of the Processor to execute. It then runs the function and waits for it to complete so it can report that the job’s computational resources can be used by other jobs.
Despite the minimal specification of Processors, there are certain responsibilities they are expected to complete. Each Processor must mark when it starts the job, when it ends the job, and whether or not the job was successful. They should also free up any disk space they utilize and if they’re successful they should remove the raw data from Temporary Storage. Since every Processor needs to do this stuff a framework is provided by the system to make it easy and to keep things DRY. The framework was designed with the goal of allowing reusable and composable functions to be created which can then be joined together into a single pipeline. Each function in the pipeline takes a job_context dictionary as an input and must output another (or the same) dictionary which can be passed into the next function of the pipeline. This behavior can be utilized by calling processors.utils.run_pipeline(), which takes two arguments: an initial job_context dictionary and a list of functions.
To illustrate this let’s take a look at the Processor function for our example:
def build_index(job_id: int) -> None:
utils.run_pipeline({"job_id": job_id},
[utils.start_job,
_set_job_prefix,
_prepare_files,
_process_gtf,
_create_index,
_zip_index,
utils.upload_processed_files,
utils.cleanup_raw_files,
utils.end_job])
Any function which returns a job_context containing ”success”: False will cause the pipeline to terminate and the job to be marked as a failure. Notice that the list of functions is composed of calls to functions defined in the utils namespace and private functions from the current namespace. The functions defined in the utils namespace take care of the common functionality I discussed in the previous paragraph. They pretty much all do what their names imply, but it will be helpful to note that in addition to marking the ProcessorJob as started, utils.start_job() also retrieves the ProcessorJob and Batch objects from the database and adds them to the job_context. So now let’s take a look at our pipeline step by step starting with:
def _set_job_prefix(job_context: Dict) -> Dict:
job_context["job_dir_prefix"] = "processor_job_" + str(job_context["job_id"])
return job_context
This adds the key “job_dir_prefix” to the job_context, which serves a similar purpose to the variable named similarly from the Downloaders System section: it makes all paths related to this job unique on the local disk. Next we run _prepare_files() which has the purpose of pulling the files from Temporary Storage and unzipping them so they’re ready to be processed.
def _prepare_files(job_context: Dict) -> Dict:
# Transcriptome index processor jobs have only one batch. Each
# batch has a fasta file and a gtf file
batch = job_context["batches"][0]
fasta_file = File.objects.get(batch=batch, raw_format__exact="fa.gz")
gtf_file = File.objects.get(batch=batch, raw_format__exact="gtf.gz")
fasta_file.download_raw_file(job_context["job_dir_prefix"])
gtf_file.download_raw_file(job_context["job_dir_prefix"])
gzipped_fasta_file_path = fasta_file.get_temp_pre_path(job_context["job_dir_prefix"])
gzipped_gtf_file_path = gtf_file.get_temp_pre_path(job_context["job_dir_prefix"])
job_context["fasta_file_path"] = gzipped_fasta_file_path.replace(".gz", "")
job_context["gtf_file_path"] = gzipped_gtf_file_path.replace(".gz", "")
with gzip.open(gzipped_fasta_file_path, "rb") as gzipped_file, \
open(job_context["fasta_file_path"], "wb") as gunzipped_file:
shutil.copyfileobj(gzipped_file, gunzipped_file)
with gzip.open(gzipped_gtf_file_path, "rb") as gzipped_file, \
open(job_context["gtf_file_path"], "wb") as gunzipped_file:
shutil.copyfileobj(gzipped_file, gunzipped_file)
job_context["fasta_file"] = fasta_file
job_context["gtf_file"] = gtf_file
return job_context
With our files unzipped locally, we’re ready to start processing by calling process_gtf(). This function cleans the genome annotation file (the gtf_file in this case) and creates a mapping from genes to transcripts for the given genome. If you’re not sure what that means, don’t worry about it; the upshot is that job_context["gtf_file_path"] is updated to point to the cleaned GTF file and the ”genes_to_transcripts_path” key is added to the job_context. Now we’re ready to create a Salmon Index using RSEM and Salmon:
def _create_index(job_context: Dict) -> Dict:
# Prepare the output directories.
work_dir = job_context["gtf_file"].get_temp_dir(job_context["job_dir_prefix"])
job_context["output_dir"] = os.path.join(work_dir, "index")
rsem_index_dir = os.path.join(work_dir, "rsem_index")
os.makedirs(rsem_index_dir, exist_ok=True)
# RSEM takes a prefix path and then all files generated by it will
# start with that
rsem_prefix = os.path.join(rsem_index_dir,
job_context["fasta_file"].get_base_name())
rsem_command_string = (
"rsem-prepare-reference --gtf {gtf_file}"
" --transcript-to-gene-map {genes_to_transcripts} {fasta_file} {rsem_prefix}"
)
rsem_formatted_command = rsem_command_string.format(
gtf_file=job_context["gtf_file_path"],
genes_to_transcripts=job_context["genes_to_transcripts_path"],
fasta_file=job_context["fasta_file_path"],
rsem_prefix=rsem_prefix
)
rsem_completed_command = subprocess.run(rsem_formatted_command.split())
salmon_command_string = ("salmon --no-version-check index -t {rsem_transcripts}"
" -i {index_dir} --type quasi -k {kmer_size}")
# This is the property we added in the Surveyor System section
# to denote whether this index is for long or short average read lengths.
kmer_size_property = BatchKeyValue.objects.get(batch=job_context["batches"][0],
key="kmer_size")
# rsem-prepare-reference outputs a transcripts.fa file which needs
# to be passed into salmon.
rsem_transcripts = rsem_prefix + ".transcripts.fa"
salmon_formatted_command = salmon_command_string.format(
rsem_transcripts=rsem_transcripts,
index_dir=job_context["output_dir"],
kmer_size=kmer_size_property.value)
salmon_completed_command = subprocess.run(salmon_formatted_command.split())
return job_context
It’s a big function, perhaps even a bit too big, but it really boils down to two shell calls. The first shell call to rsem-prepare-referenceoutputs the rsem_transcripts file which is then passed into salmon index. salmon-index outputs an entire directory which we want all of.
The next call in the pipeline, _zip_index(), takes care of zipping the entire directory into a single tarball along with setting job_context["files_to_upload"] = [job_context["gtf_file"]]. The reason for setting that property is that utils.upload_processed_files will look for it and, if it’s present in job_context, will only upload the files in that list to Permanent Storage. The File.upload_processed_file() method will use the File’s processed_format property to determine which file should be uploaded. Therefore even though we don’t want to upload the gtf_file, that File object knows how to upload its processed file, which in this case is the tarball we create in _zip_index(). All that’s left in our pipeline is utils.cleanup_raw_files() which will clear the raw data out of Temporary Storage and utils.end_job() which will free up the local disk space for the next job and mark the job as a success.
6. Conclusion
Once the ProcessorJob has been marked as a success in the database, we’re done with our example! We’ve certainly skipped over some sticky parts such as dealing with the idiosyncrasies of Ensembl’s REST API or less interesting code such as failure logging and handling. However, the goal of this blog post was to illustrate how the different components of the system work together to actually get some data from The Internet™ and refine it. I also only barely mentioned the Foreman, which makes sure that when failure inevitably happens, jobs will be retried a couple times before an administrator of the system is notified.
I’ve also omitted a discussion of the Aggregator, but that’s mostly because it’s still being fully specced out. What is defined about it is that it will have a REST API which our various interfaces will use to search, aggregate, and download data stored in refine.bio’s compendium. We’re currently gathering user feedback to better define its functionality. We’re also currently working on finding additional team members to help us implement it.

